1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
|
#!/usr/bin/env python3
"""
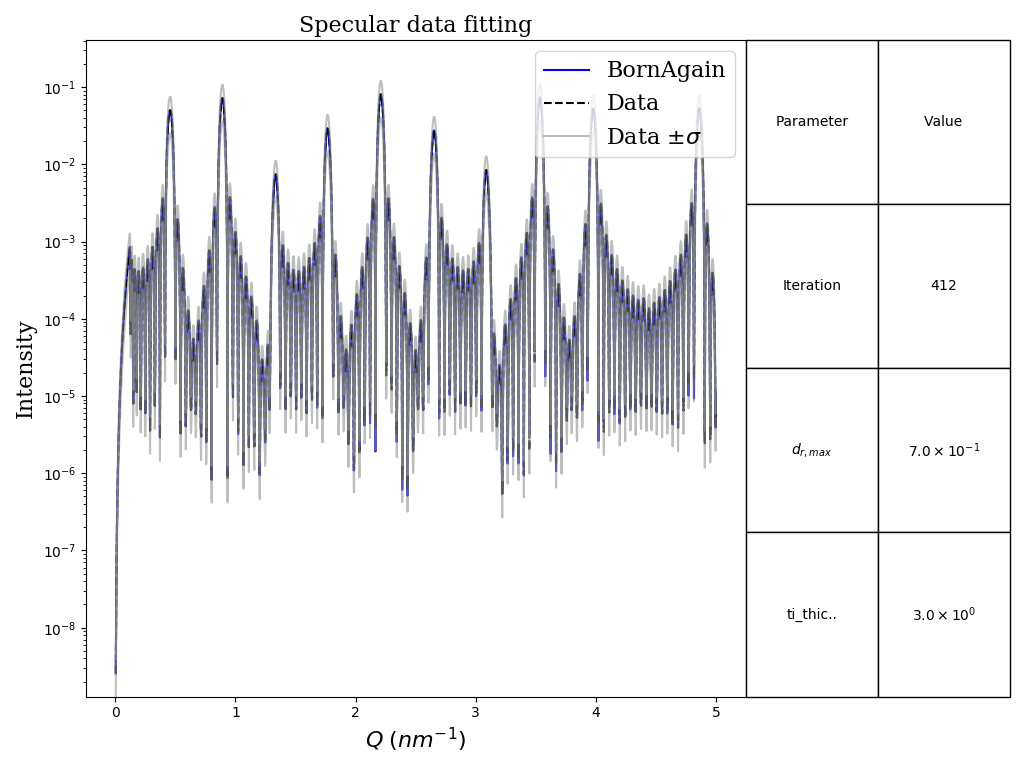
Example demonstrates how to fit specular data.
Our sample represents twenty interchanging layers of Ti and Ni. We will fit
thicknesses of all Ti layers, assuming them being equal.
Reference data was generated with GENX for ti layers' thicknesses equal to 3 nm
This example uses exactly the same data as in FitSpecularBasics. However this
time we will add artificial uncertainties and use RQ^4 view.
Besides we will set Chi squared with L1-normalization as the objective metric
and use genetic algorithm as the minimizer.
"""
import os
import numpy as np
from matplotlib import pyplot as plt
import bornagain as ba
from bornagain import ba_fitmonitor
import FitSpecularBasics as fsb
def run_fitting():
"""
Setup simulation and fit
"""
real_data = fsb.get_real_data_values()
# setting artificial uncertainties (uncertainty sigma equals a half
# of experimental data value)
uncertainties = real_data*0.5
fit_objective = ba.FitObjective()
fit_objective.addSimulationAndData(fsb.get_simulation, real_data,
uncertainties)
plot_observer = ba_fitmonitor.PlotterSpecular(units=ba.Coords_RQ4)
fit_objective.initPrint(10)
fit_objective.initPlot(10, plot_observer)
fit_objective.setObjectiveMetric("Chi2", "L1")
params = ba.Parameters()
params.add("ti_thickness",
50*ba.angstrom,
min=10*ba.angstrom,
max=60*ba.angstrom)
minimizer = ba.Minimizer()
minimizer.setMinimizer("Genetic", "", "MaxIterations=40;PopSize=10")
result = minimizer.minimize(fit_objective.evaluate, params)
fit_objective.finalize(result)
if __name__ == '__main__':
datadir = os.getenv('BA_EXAMPLE_DATA_DIR', '')
data_fname = os.path.join(datadir, "genx_interchanging_layers.dat.gz")
fsb.get_real_data(data_fname)
run_fitting()
plt.show()
|
 BornAgain
≻ 20
≻ Documentation ≻ Python scripting ≻ Fitting ≻ Extended examples ≻ Considering uncertainties
BornAgain
≻ 20
≻ Documentation ≻ Python scripting ≻ Fitting ≻ Extended examples ≻ Considering uncertainties